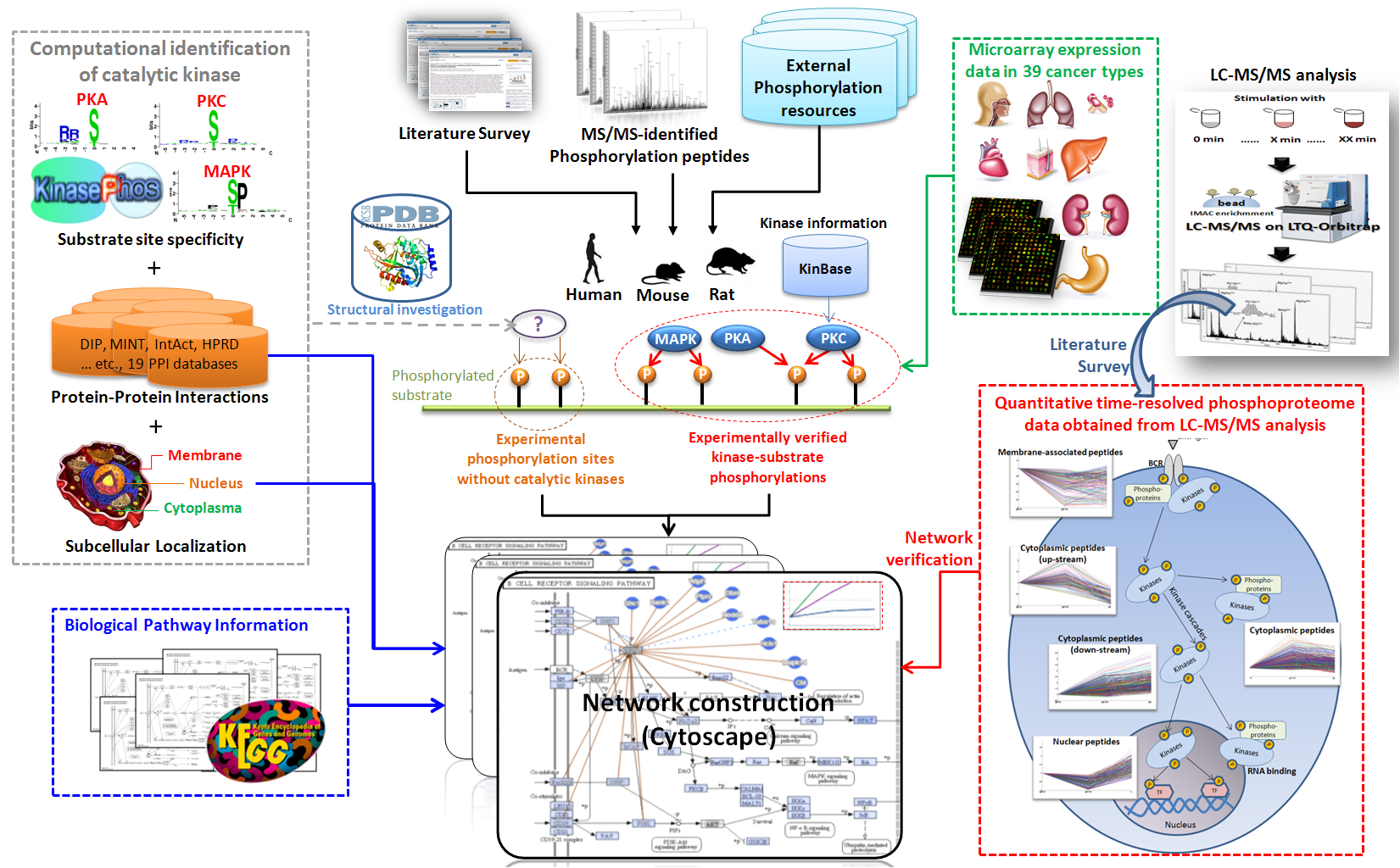
Protein phosphorylation catalyzed by kinases plays crucial roles in regulating a variety of intracellular processes. Due to an increasing number of in vivo phosphorylation sites that has been identified by mass spectrometry (MS)-based proteomics, the RegPhos was developed to explore protein phosphorylation networks in human. In this update, we not only enhance the data content in human but also investigate kinase-substrate phosphorylation networks in mouse and rat. The experimentally validated phosphorylation sites as well as their catalytic kinases were extracted from public resources and MS/MS phosphopeptides were manually curated from research articles. RegPhos 2.0 aims to provide a more comprehensive view of intracellular signaling networks by integrating the information of metabolic pathways and protein-protein interactions. Additionally, analyzing the phosphoproteome profile of time-dependent cell activation that was obtained from LC-MS/MS analysis, we deciphered not only the consistent scheme in B cell receptor (BCR) signaling pathway but also novel regulating molecules that may involve in it. In addition, to help users efficiently identify the candidate biomarkers in cancers, 30 microarray experiments, including 39 cancerous versus normal cells, were analyzed for detecting cancer-specific expressed genes coding for kinases and their substrates. Furthermore, this update features an improved web interface to facilitate convenient access to the structural environment of phosphorylation sites on protein tertiary structures.